Time for some probing questions
15 May 2018 by Evoluted New Media
Biomedical research is heavily dependent on utilising biological and chemical probes to understand the functionality of medically relevant proteins. Yet, when it comes to chemical probes, we aren’t being careful enough to validate our findings – and this is contributing to the reproducibility crisis. But help is at hand…
Understanding fundamental mechanisms in biology, uncovering processes causing disease and validating targets for drug discovery are key goals of biomedical research. Scientists employ chemical and biological tools to tackle these goals.1 If selected and applied carefully, RNA interference and CRISPR can be used as robust biological tools to study the function of proteins and genes. The same rigour is required for chemical probes.
Small-molecule chemical tools, or probes, are widely-used in mechanistic studies and can provide evidence that the activity of a protein of interest is important in a biological or pathological process – and also that this activity can be modified pharmacologically, thus setting a firm foundation for drug discovery.2 As both biological and chemical tools have limitations, the use of the two in parallel is recommended where possible.1 Furthermore, it is imperative that scientists using these tools make strong efforts to verify that they are of appropriate high-quality, and must understand their limitations. Failure to do so risks producing misleading results and serious errors in the biomedical literature, contributing to the worrying lack of reproducibility and robustness in biomedical research and the selection of inappropriate drug targets.3
Not all claimed chemical probes are useful A chemical compound must possess some basic essential properties or ‘fitness factors’ if it is to be used as an acceptable reagent with which to investigate the function of a specific protein or to validate a target in a therapeutic context.2 Compounds that possess such characteristics can be classed as high-quality chemical probes. The discovery of a valuable chemical probe for a new target can have a very positive impact, and may even unlock a new field of study, particularly if the probe is made publicly available.
For example, the discovery of JQ1 and I-BET has had a profound positive impact, leading to the uncovering of BET bromodomain biology and follow-up drug discovery.4 However, the use of flawed chemical probes has had a profound negative impact on biomedical research, distracting attention away from legitimate drug targets and wasting time and resources. One such example of a flawed probe is the natural product epigallocatechin-3-gallate, which is known to interfere with assay detection methods, and yet has been used to study and validate the DYRK1A kinase as a drug target.4–6 There are countless other examples
To be useful in the study of a complex biological system, a chemical probe must bind or inhibit a target of interest with sufficient potency to modulate its activity at a biologically appropriate concentration. A chemical probe should also have the ability to permeate living cells and be sufficiently selective to build confidence that the observed effects are due to modulation of the intended molecular target. Potency, selectivity and permeability are therefore key fundamental properties of chemical probes.
While these three features are key, there are additional fitness factors that should also be considered when assessing if a chemical probe is fit-for-purpose2. Table 1 details the fitness factors that should be considered when choosing a chemical probe, grouping them into four broad categories2: namely, chemistry, potency, selectivity and context.
Within the chemistry category, it is worth noting that chemical reactivity commonly drives promiscuous binding. Several computational methods have been developed to alert scientists to chemical substructures that can cause chemical reactivity. Other methods such as ‘Badapple’ have also proved valuable, alerting scientists to possible promiscuous probe binding irrespective of chemical reactivity.7 Moreover, filters that identify potential pan-assay interference compounds (PAINS)6 are useful and can identify substructures of compounds that are associated with an increased risk of interference with assay detection methods. However, we recommend that, wherever possible, the chemical reactivity, promiscuity, aggregation and pan-assay interference potential of a probe are tested in the lab, since the computational methods used to generate substructural alerts can predict false positives.
Avoiding the use of reactive, promiscuous or assay-interfering compounds is essential to prevent cases such as resveratrol, which appeared to revolutionise the study of sirtuins in aging until it was discovered that this compound was actually interfering with the assay detection method.4 Validating the identity and purity of the chemical probe under consideration is also highly recommended, even when the chemical probe is sourced from a reputable supplier.
Potency should be measured in biochemical and cell-based assays and using different ‘orthogonal’ methods where possible. Typically, a good probe will be active at concentrations below 100 nM in biochemical assays and at concentrations no higher than 1–10 uM in cellular assays. Moreover, chemically similar structural analogues with varying potencies, including closely related inactive analogues, are powerful tools for deciphering mechanisms. Such analogues enable the study of structure-activity relationships (SAR) for both biochemical and cellular effects which when in agreement help build confidence that the biological consequences are due to modulation of the target under study.
It is also vital to determine the selectivity of chemical probes in order to ensure that their observed effects are not the result of off-target binding to other proteins. To be fit-for-purpose a probe must be sufficiently selective for its target protein over other members of the same protein family in biochemical assays, as well as proteins from other families. The validation of this selectivity in a cellular context is highly advisable. Inactive or less active analogues can be used as controls in such cell-based assays, to give confidence that the observed effects are the result of on-target binding. We also highly recommended the use of a second chemical probe of a different class or ‘chemotype’. Chemical probes from different chemotypes are less likely to bind the same off-targets, increasing confidence that the observed phenotypic effects result from on-target protein binding.
Finally, all of these fitness factors have to be considered in the appropriate biological context (Table 1, category 4). Genetic methods and biological tools should be used whenever possible to cross-validate or otherwise the effects observed using chemical probes. Rescue of the effect of a chemical probe using a probe-resistant form of the target is a powerful control and absence of activity in cells lacking the target can be valuable biological controls. Extensive assessment of chemical probes in many different cellular and animal models will build further confidence – but note that probes for use in animals such as rodents are required to have appropriate pharmacokinetic properties. Many researchers do not have access to custom chemical synthesis in their laboratories. Therefore, improved commercial availability of good chemical probes is essential.
A critical determinant in the interpretation of the effects of a chemical probe in a cellular or organismal context is the provision of clear evidence of the intended on-target activity in the biological model using appropriate biomarkers that clearly and ideally quantitatively demonstrate target engagement and modulation of the downstream biochemical events. We strongly caution against increasing the concentration or dose of a chemical probe progressively until a desired phenotype, especially cell death, is achieved – and then attributing this effect to modulation of the target of interest. Evidence using on-target binding/modulation for the probe is needed and ideally also biomarker data informing on effects on known or suspected off-targets. Proteome-wide methods are now available for compound profiling in cells.
Overall, there are many properties to consider before selecting a chemical probe for a biomedical experiment. On the other hand, overly prescriptive thresholds for these fitness factors could stifle progress. Ideal probes may not always be available for the initial investigation of a given protein and early prototype compounds can act as pathfinders and enable progress. The perceived fitness of a chemical probe can evolve over time, as it becomes better characterised by the wider scientific community. Importantly, early chemical probes are often superseded by better probes as a field of study progresses. Compromises on properties may be necessary at any given time, but an understanding of the limitations of the probes used is essential.2 Accordingly, we highly recommended very careful consideration and assessment of the fitness factors before selecting the most suitable up-to-date chemical probes for biological experimentation.1,2
The perils of choosing a flawed chemical tool Despite the existence of many reviews and perspectives detailing the desired and unacceptable properties of chemical probes,1,2 their selection for biomedical experiments is prone to historical and commercial biases. Scientists commonly select chemical probes using web-based search engines or based on earlier well-cited literature reports.4 Both strategies are heavily biased toward older (and often flawed) probes. Another common practice is to use vendor catalogues for chemical probe selection. However, vendor catalogues may not contain the most up-to-date information and often do not discriminate between low and high quality chemical probes. Accordingly, many biomedical studies use suboptimal or flawed chemical probes, data are misinterpreted and the scientific literature becomes populated with erroneous results, constituting an enormous waste of time and resources within the scientific community amounting to billions of dollars.
The historical PI3 kinase inhibitor LY294002 is an excellent example of a chemical probe that has been used inappropriately. It was developed in the 1990s and at that time it was (alongside the potent but reactive and unstable natural product wortmannin) the best chemical probe available with which to study the function of PI3 kinases.1 However, LY294002 has only modest biochemical potency (IC50 = 1.6 ?M) against PI3 kinases and later research revealed its lack of selectivity for PI3 kinase – with effects not only against numerous lipid and protein kinases, but also members of other protein families such as bromodomains.1 Today, there are several selective PI3 kinase inhibitors which are far more potent than LY294002, such as PI-103 and the clinical candidate pictilisib (GDC-0941). Yet despite the existence of these better chemical probes, LY294002 continues to be widely used in biomedical research, appearing in more than 1000 publications in 2016.1 Such publications will likely contribute further to the ongoing use of this now outdated chemical probe in new studies. Moreover, LY294002 is still sold by many compound vendors as a PI3 kinase inhibitor, without alerting buyers to its lack of selectivity and unsuitabilty. This example illustrates the limitations of the most common approaches to chemical probe selection.
Unwise choices can lead to enormous overall cost, both in terms of time spent on biomedical research and the integrity of published data.
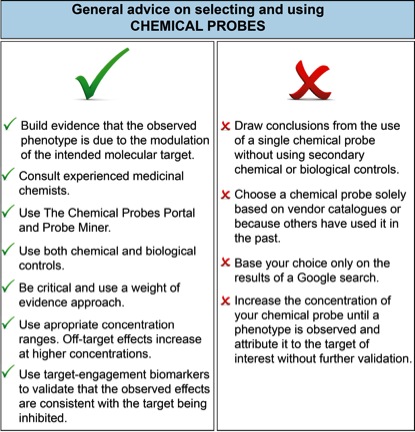
Figure 1 lists user-friendly do’s and don’ts for chemical probe selection. It is clear that biomedical researchers, scientific reviewers and journal editors urgently need better resources to help them assess and select chemical probes for biomedical experiments.
The Chemical Probes Portal In 2015, The Chemical Probes Portal (http://www.chemicalprobes.org/) was launched as a non-profit community-driven expert-curated resource to aid selection of best-available chemical tools and advise on current best practice for use of chemical probes in biomedical research and target validation.5 The Chemical Probes Portal offers manually collated and curated information about chemical probes provided by expert reviewers. It grades selected chemical probes using a star-based scoring system, as well as providing informative comments relating to probes for specific protein targets. Today, the portal covers more than 400 chemical probes, all published, and each of them subject to subsequent expert peer review and accompanied by an inventory of relevant properties, as well as guidance on appropriate use. The Portal also has information and warnings about ‘historical compounds’, now superseded by others, to discourage the misapplication of these reagents as if they were high-quality chemical probes useful today.
In the past three years, The Chemical Probes Portal has had a marked impact on improving chemical probe selection and use. It is accessed by ca 7000 users per month.1 However, it is extremely challenging to provide expert reviews of all of the compounds that could potentially be used as chemical probes. Each expert review is an extensive, manual effort based on an in-depth review in the context of an understanding of the literature and the field. Accordingly, we felt there was a need for an additional, complementary resource that could exploit large-scale publicly available medicinal chemistry and chemical biology datasets to objectively assess and prioritise chemical probes for further assessment and use.
Objective, quantitative, data-driven assessment To assess how many of the compounds listed in public databases could be useful as chemical probes, we recently analysed large-scale medicinal chemistry and pharmacological data sets integrated into our canSAR knowledgebase (http://cansar.icr.ac.uk/).8 We established key criteria that define minimum levels of potency, selectivity and permeability which – while not a guarantee that a chemical tool will make a useful probe – we believe all chemical probes should be required to satisfy.
Surprisingly, we found that the data on chemical tool selectivity held in public medicinal chemistry databases is alarmingly limited and most compounds failed to meet our probe criteria. Moreover, we found that chemical tools that did fulfil our minimum requirements for potency, selectivity and permeability are available only for an unacceptably low percentage (1.2%) of the human proteome. Accordingly, it is imperative to improve the characterisation of available chemical probes across multiple targets – especially in terms of selectivity – alongside the ongoing discovery of chemical probes for new targets. To provide the available information and unbiased assessment of chemical probes, complementary to The Chemical Probes Portal, we decided to create a public user-friendly website resource to democratise access to the most up-to-date information on potential chemical tools.
The Probe Miner online resource We developed Probe Miner (http://probeminer.icr.ac.uk/) as a non-profit, large-scale, data-driven and regularly updated resource to enable the unbiased objective assessment of chemical probes for specific biological targets.8 We developed six different chemical probe scoring metrics based on fitness factors2 of potency, selectivity, permeability, SAR, inactive analogues and PAINS and used these to provide an overall ‘global score’ for any given probe.
Using this approach we analysed >3.9 million experimental activities for >1.8 million compounds available in public medicinal chemistry databases, providing an objective and systematic scoring of 355,305 chemical tools against 2,220 human protein targets.8
Probe Miner is designed to be searched by biological target. After choosing a target of interest, the resource directs users to an interactive graphical overview page that summarises the quality of existing chemical tools for the selected target and provides an easy-to-navigate distribution of the top-ranking chemical probes, plus a chemical probe viewer summarising the scores. The user is able if they wish to take a deep-dive into the data before selecting a chemical probe – in this way they can access a more detailed page describing the compound and the target of interest, with links to the original data. Although a default setting is provided for ease of use, advanced users can customise the weighting of each score (potency, selectivity, permeability, SAR, inactive analogues and PAINS) within the global score to facilitate a custom ranking of the probes. Finally, Probe Miner also enables users to access and download all of the underlying data.
We found that there is quite a high degree of consistency between the probes ranked highly by Probe MinerTM and those recommended by experts on the Chemical Probes Portal. Specifically, 67% of the probes highly rated by experts also score highly using the objective assessment methodology of Probe Miner.8 However, no one method is perfect, both resources have their own benefits and limitations, and some differences in probe rankings are clearly seen. By its nature, Probe Miner captures data from public medicinal chemistry databases which may lack some key chemical biology data published in non-medicinal chemistry journals. Database errors are of course inevitable and it is incredibly challenging to identify all of these. Thus we invite the research community to inform us of any database errors so that we can find appropriate solutions and continue to improve the performance of Probe Miner (email: chemprobes@icr.ac.uk).
An advantage of Probe Miner is its objectivity and its much broader coverage of the proteome than the manually-curated Portal. We highly recommended using Probe Miner alongside The Chemical Probes Portal whenever possible, to benefit from the complementary and synergistic benefits that each resource provides. Examples are provided in the publication.8 For ease, we provide one-click links for individual probes from Probe Miner to The Chemical Probes Portal.
Importantly, Probe MinerTM is very efficient at discouraging the use of suboptimal or flawed probes when better options are available, such as in the case of PI3KCA for which the suboptimal chemical tool LY294002 does not appear among the top scoring probes whereas potent and selective inhibitors like pictilisib (GDC-0941) do appear.
For new users, we provide an online tutorial video to facilitate navigation of the Probe Miner resource (http://probeminer.icr.ac.uk/#/tutorial).
Next steps Overall, we feel that Probe Miner is a very valuable addition to the toolbox for chemical probe prioritisation and has been already accessed by more than 1,200 users since we released the first beta-version in July 2017. We will maintain Probe Miner and provide regular automatic updates that ensure its topicality. By curating missing chemical biology data into Probe Miner we also plan to increase its coverage of compounds. Probe Miner will facilitate the identification of potentially useful probes for expert review at the complementary Chemical Probes Portal. We keep in close contact with users so that we can continuously improve the Probe Miner user interface and the information available, ensuring that the research community reaps the maximum benefit. We hope that, when used together with The Chemical probes Portal, Probe Miner will promote best practice in chemical biology and thereby help to overcome the commonly poor selection and use of chemical probes and the alarming lack of selectivity of many of them. In turn, this should contribute to improving data reproducibility and robustness in biomedical research and enhancing the selection of the best targets for drug discovery.
[box type="info" ]Probe Miner
Probe Miner is designed to be searched by biological target. After choosing a target of interest, the resource directs users to an interactive graphical overview page that summarises the quality of existing chemical tools for the selected target and provides an easy-to-navigate distribution of the top-ranking chemical probes, plus a chemical probe viewer summarising the scores. The user is able if they wish to take a deep-dive into the data before selecting a chemical probe – in this way they can access a more detailed page describing the compound and the target of interest, with links to the original data. Although a default setting is provided for ease of use, advanced users can customise the weighting of each score (potency, selectivity, permeability, SAR, inactive analogues and PAINS) within the global score to facilitate a custom ranking of the probes. Finally, Probe Miner also enables users to access and download all of the underlying data. [/box]
AUTHORS

Albert A. Antolin, Paul Workman and Bissan Al-Lazikani (L-R) are based at the Cancer Research UK Cancer Therapeutics Unit and the Department of Data Science at The Institute of Cancer Research in London.
Paul Workman is also Board Director of the non-profit Chemical Probes Portal; all the present authors are also authors on the paper describing Probe Miner.
REFERENCES
- Blagg, J. & Workman, P. Choose and Use Your Chemical Probe Wisely to Explore Cancer Biology. Cancer Cell 32, 9–25 (2017).
- Workman, P. & Collins, I. Probing the Probes: Fitness Factors For Small Molecule Tools. Chem. Biol. 17, 561–577 (2010).
- Begley, C. G., Buchan, A. M. & Dirnagl, U. Robust research: Institutions must do their part for reproducibility. Nature 525, 25–27 (2015).
- Arrowsmith, C. H. et al. The promise and peril of chemical probes. Nat. Chem. Biol. 11, 536–541 (2015).
- De la Torre, R. et al. Epigallocatechin-3-gallate, a DYRK1A inhibitor, rescues cognitive deficits in Down syndrome mouse models and in humans. Mol. Nutr. Food Res. 58, 278–288 (2014).
- Baell, J. & Walters, M. A. Chemical con artists foil drug discovery. Nature 513, 481–3 (2014).
- Yang, J. J. et al. Badapple: Promiscuity patterns from noisy evidence. J. Cheminform. 8, 1–14 (2016).
- Antolin, A. A. et al. Objective , Quantitative , Data-Driven Assessment of Chemical Probes. Cell Chem. Biol. 1–12 (2018).